Biological Membranes
Introduction
Biological membranes are comprised of amphiphilic lipids forming a fluid bilayer that serves as a two-dimensional interface where a number of associated proteins perform a variety of functions. While each kind of 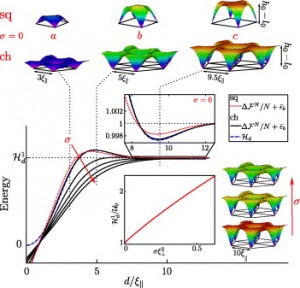 protein is present at a relatively low concentration, the sheer number of protein types makes the cellular membrane a very crowded environment. However, many cell functions require the active and passive recruitment of only certain proteins to specific regions on the cell membrane, in a very poorly understood process. Eventually, proteins localize, make macromolecular complexes and may become anchored to larger structures on the inside or the outside of the cell, thereby becoming effectively immobile for an appreciable time. The consequence of this immobilisation or “slowing-down” is that the complexes themselves compartmentalize the membrane and act as a scaffold structure for the dynamics of other proteins or even the lipids themselves. Furthermore, the accumulation of proteins is often connected to the change of the underlying lipid matrix, resulting in locally variable elastic properties of the system. These variations may have a significant impact on the ability of the proteins to mutually interact, simply because the formation of molecular bonds couples to the elasticity of the membrane. The complexation both exerts and is subject to stresses from the environment. The latter are very noisy and out of equilibrium, due to a number of competing processes which occur simultaneously. This results in fluctuations, which in turn impact the protein complexation and may promote cooperative behaviour.
protein is present at a relatively low concentration, the sheer number of protein types makes the cellular membrane a very crowded environment. However, many cell functions require the active and passive recruitment of only certain proteins to specific regions on the cell membrane, in a very poorly understood process. Eventually, proteins localize, make macromolecular complexes and may become anchored to larger structures on the inside or the outside of the cell, thereby becoming effectively immobile for an appreciable time. The consequence of this immobilisation or “slowing-down” is that the complexes themselves compartmentalize the membrane and act as a scaffold structure for the dynamics of other proteins or even the lipids themselves. Furthermore, the accumulation of proteins is often connected to the change of the underlying lipid matrix, resulting in locally variable elastic properties of the system. These variations may have a significant impact on the ability of the proteins to mutually interact, simply because the formation of molecular bonds couples to the elasticity of the membrane. The complexation both exerts and is subject to stresses from the environment. The latter are very noisy and out of equilibrium, due to a number of competing processes which occur simultaneously. This results in fluctuations, which in turn impact the protein complexation and may promote cooperative behaviour.
Our research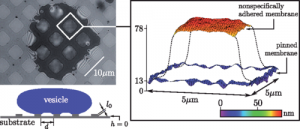 We focus on modelling specific and non-specific membrane-substrate and intra-membrane interactions which compartmentalize the membrane. We combine analytic and stochastic modelling with Langevin and effective Monte Carlo simulations to study the effects of membrane elasticity and fluctuations on the affinity of binding. Furthermore, we address the dynamics of the formation of bond clusters. We explore the interplay between the membrane environment, the mobility of proteins, and the capacity to form bonds, in the context of adhesion and protein organization in membranes.
We focus on modelling specific and non-specific membrane-substrate and intra-membrane interactions which compartmentalize the membrane. We combine analytic and stochastic modelling with Langevin and effective Monte Carlo simulations to study the effects of membrane elasticity and fluctuations on the affinity of binding. Furthermore, we address the dynamics of the formation of bond clusters. We explore the interplay between the membrane environment, the mobility of proteins, and the capacity to form bonds, in the context of adhesion and protein organization in membranes.
Collaborations
This project is developed in a tight experimental collaboration with K. Sengupta (INSERM, Marseille, FR), R. Merkel (FC Jülich, DE), S. Fenz (Univ. Wurzburg, DE) and E. Sackmann (TU München, DE), while cross-fertilization of theoretical ideas successfully takes place with the group of U. Seifert (Univ. Stuttgart, DE).
People

- We establish several methods for determining the tension and the potential strength in non-specifically adhered membranes.
Physics of Cell Adhesion: Some Lessons from Cell-mimetic Systems
- In this review we connect the observations from cell-mimetic systems with the regulation mechanisms in processes involving cell adhesion.
Nucleation of Ligand-Receptor Domains in Membrane Adhesion
- Here we predict the critical size of a nanodomain and the respective nucleation time depending on the affinity of ligand-receptor bonds.
Coexistence of dilute and densely packed domains of ligand-receptor bonds in membrane adhesion
- By analytic calculations we show that fluctuations and membrane elasticity may stabilize complex arrangements of bonds between interacting membranes. We also predict the confinement-induced renormalization of the affinity for ligand-receptor binding.
Switching from ultra-weak to strong adhesion
- Experimentally and in simulations we show that even very strong bonds can be destabilized by membrane fluctuations.
Available projects
Guided transport of diffusive particles
Coating of colloids with different sizes
Funding 

